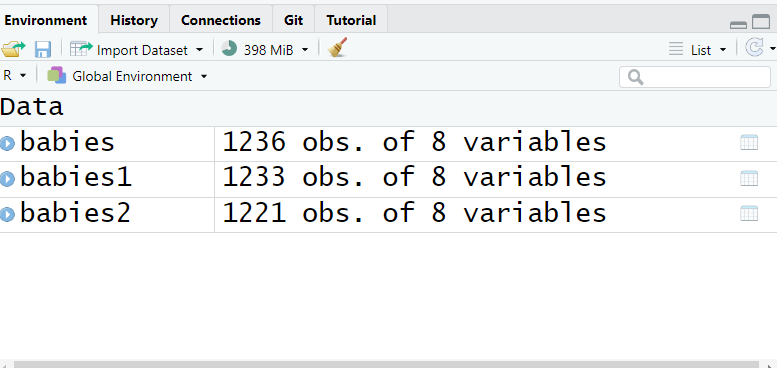
Rows: 1,236
Columns: 8
$ case <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 1…
$ bwt <int> 120, 113, 128, 123, 108, 136, 138, 132, 120, 143, 140, 144, …
$ gestation <int> 284, 282, 279, NA, 282, 286, 244, 245, 289, 299, 351, 282, 2…
$ parity <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
$ age <int> 27, 33, 28, 36, 23, 25, 33, 23, 25, 30, 27, 32, 23, 36, 30, …
$ height <int> 62, 64, 64, 69, 67, 62, 62, 65, 62, 66, 68, 64, 63, 61, 63, …
$ weight <int> 100, 135, 115, 190, 125, 93, 178, 140, 125, 136, 120, 124, 1…
$ smoke <int> 0, 0, 1, 0, 1, 0, 0, 0, 0, 1, 0, 1, 1, 1, 0, 0, 1, 1, 0, 1, …Correlation & Least Squares Regression
Week 9, Lecture 3 Lab 7
December 3th, 2024
Correlation
Let’s Load This Week’s Data Set
Correlation Coefficient
The correlation coefficient is rarely computed by hand so we will use the cor() function to obtain our observed sample correlation coefficient.
Direction of the relationship: Positive Relationship
Outliers
Outliers can distort regression analysis in two ways:
- by inflating \(s_e\) and reducing correlation; and
- by unduly influencing the regression line.
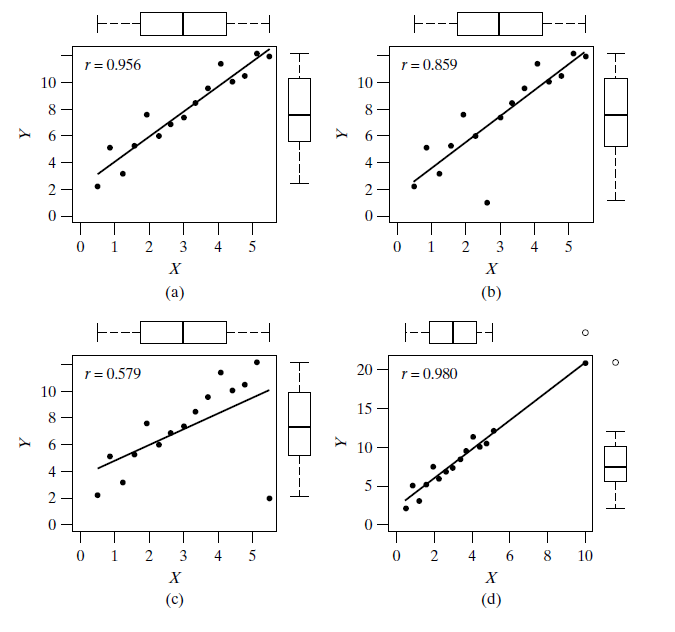
Outliers
- Leverage points are points that have the potential to greatly influence the slope of the fitted regression model.
- The further a point’s X value is from the center of the X distribution, the more leverage that point has on the overall regression model.
- Plots (c) and (d) display examples of leverage points.
- In plot (c) the leverage point is shown to actually exert its leverage on the line, tipping the regression from the bulk of the data.
- A point that has a large effect on the regression model is called an influential point.
- Plot (d) shows a leverage point (because of the extreme X value) that is not influential because the regression line does not get pulled away from the trend in the bulk of the data.
- Note that the outlier in plot (b) is not considered a leverage point—its ability to affect the slope of the line is weak as its X value is near the center of the X distribution.
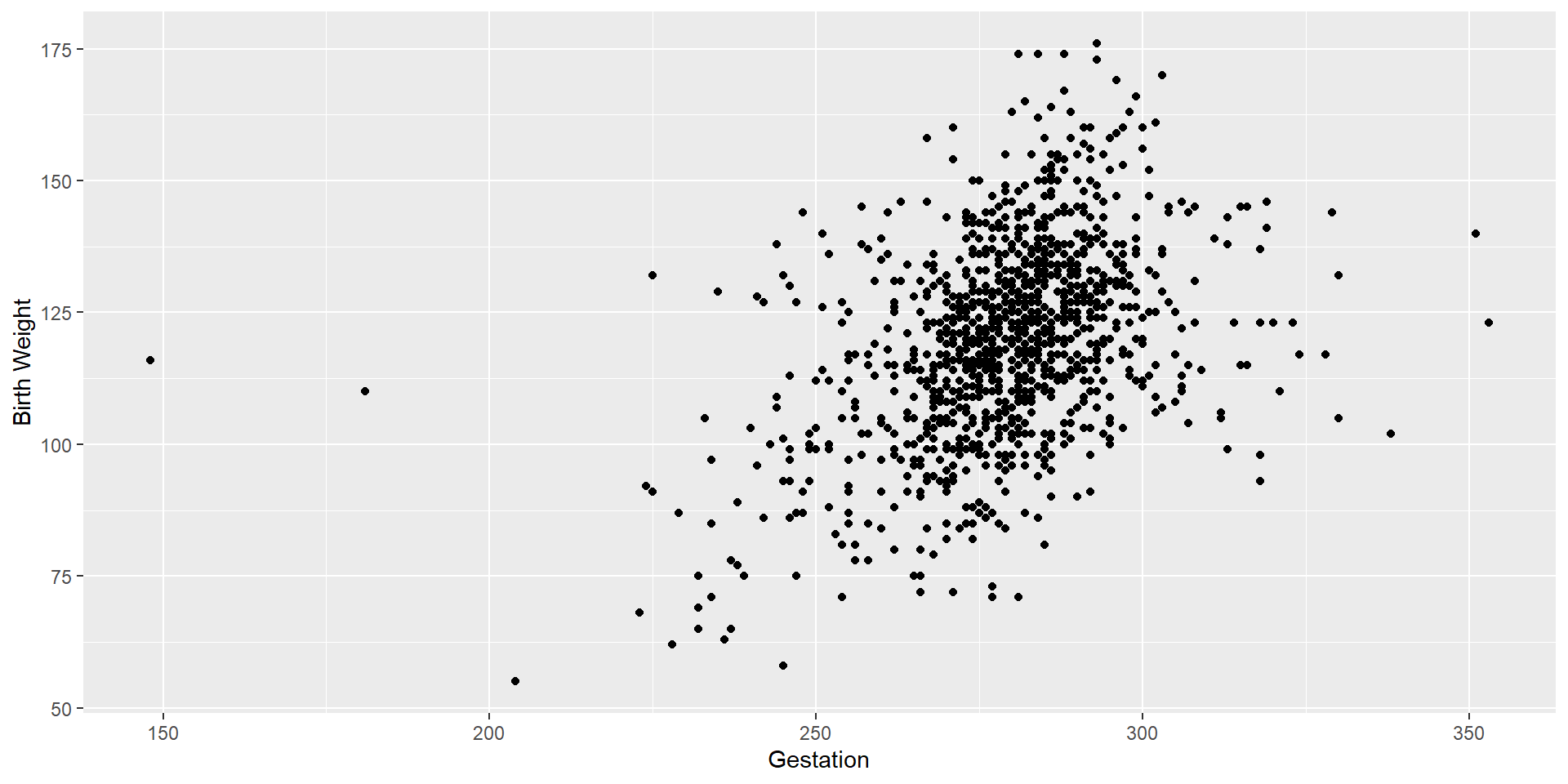
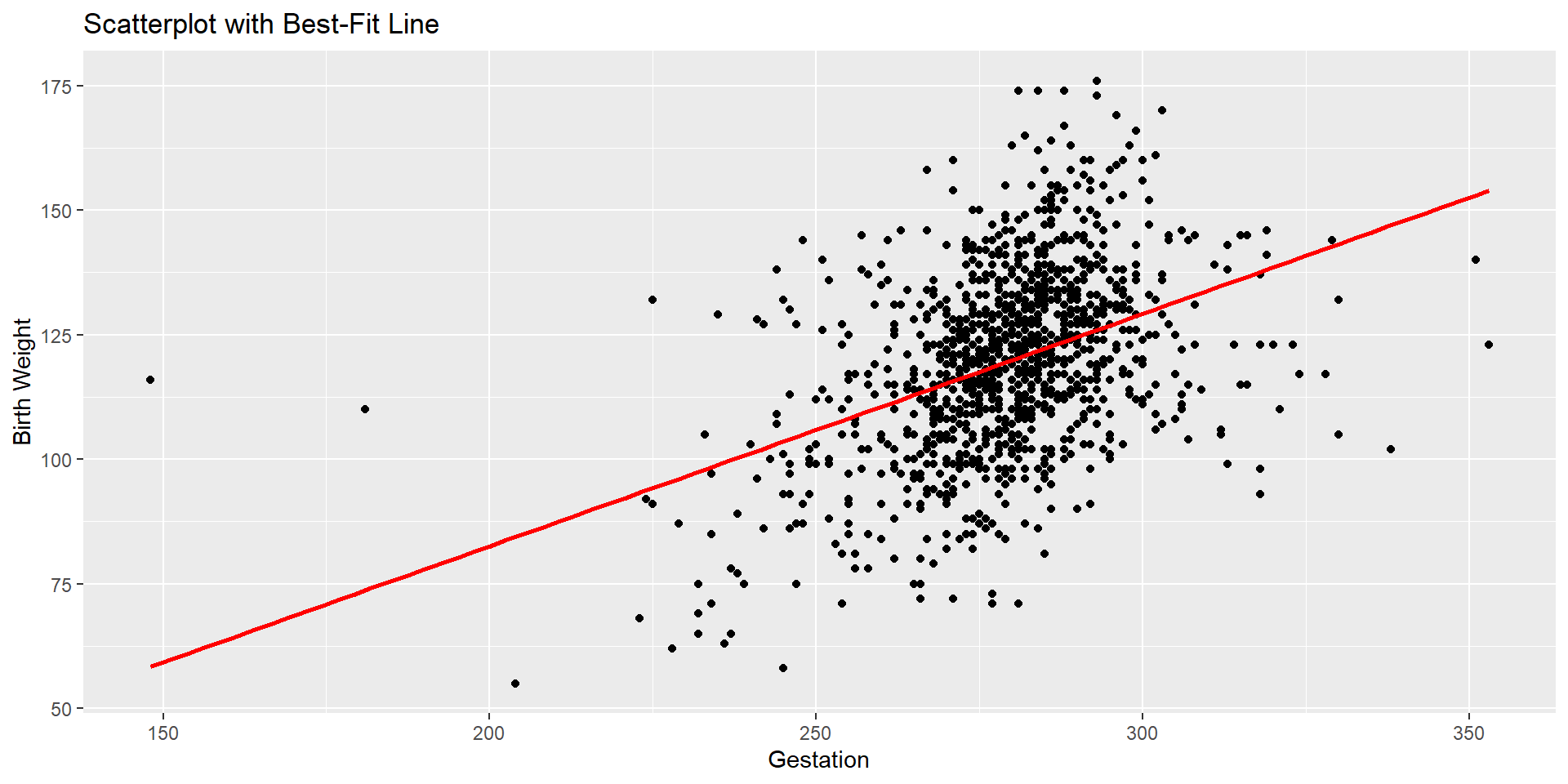
Scatterplot
Let’s see the scatterplot to look for potential outliers.
Scatterplot
Let’s see the scatterplot to look for potential outliers.
Let’s clean the data set
We see two outliers on the left-hand side of the scatterplot. Let’s remove those outliers.
Check your environment pane and notice that the number of observations is decreased.
Let’s see the changes
Let’s check the correlation coefficient and scatterplot again.
Let’s see the changes
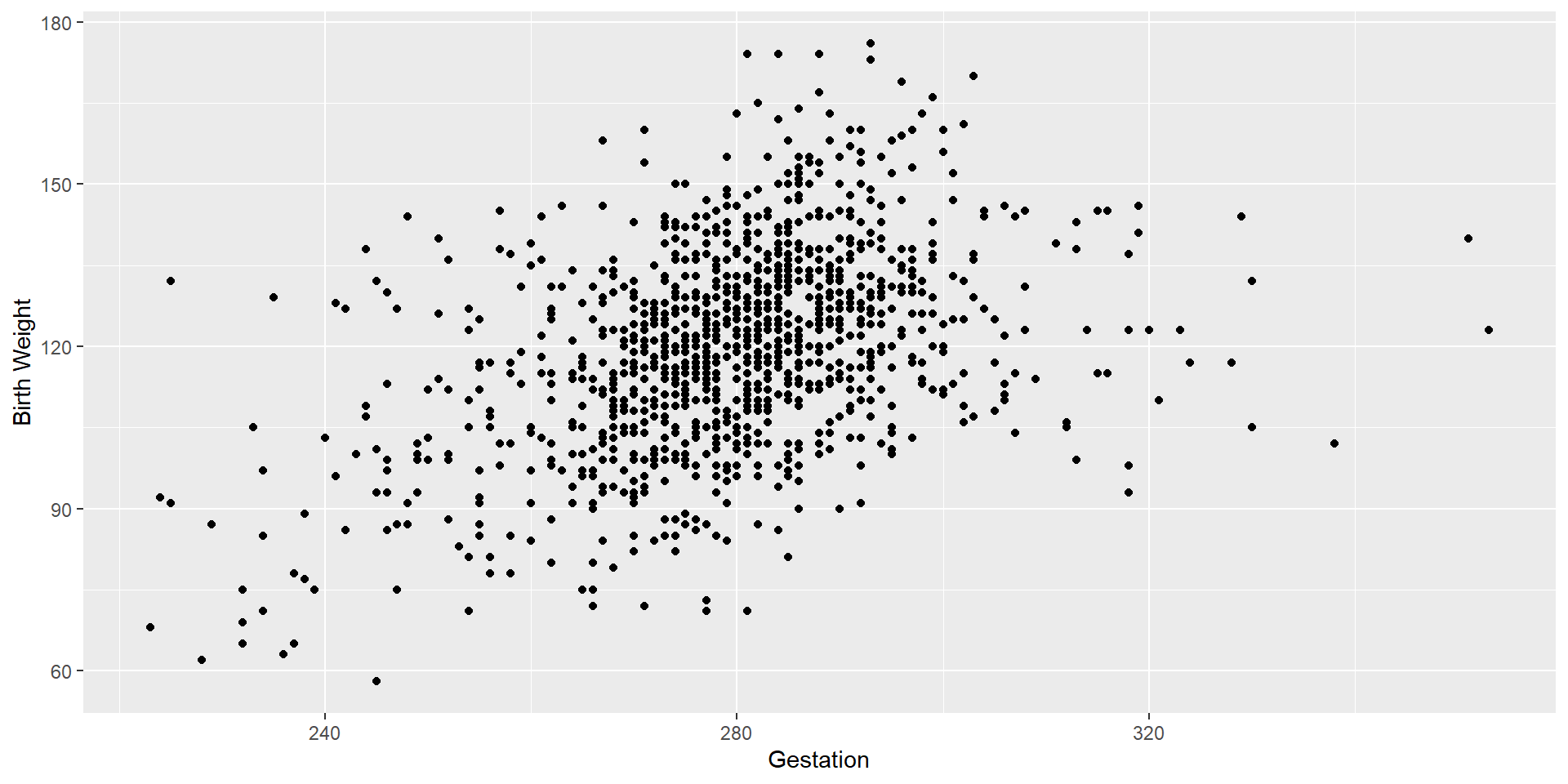
We see some potential outliers on the right-hand side of the scatterplot. Let’s remove those as well.
Let’s clean the data set one more time
Let’s see the changes
Let’s check one last time and decide if we really need to remove outliers.
Let’s see the changes
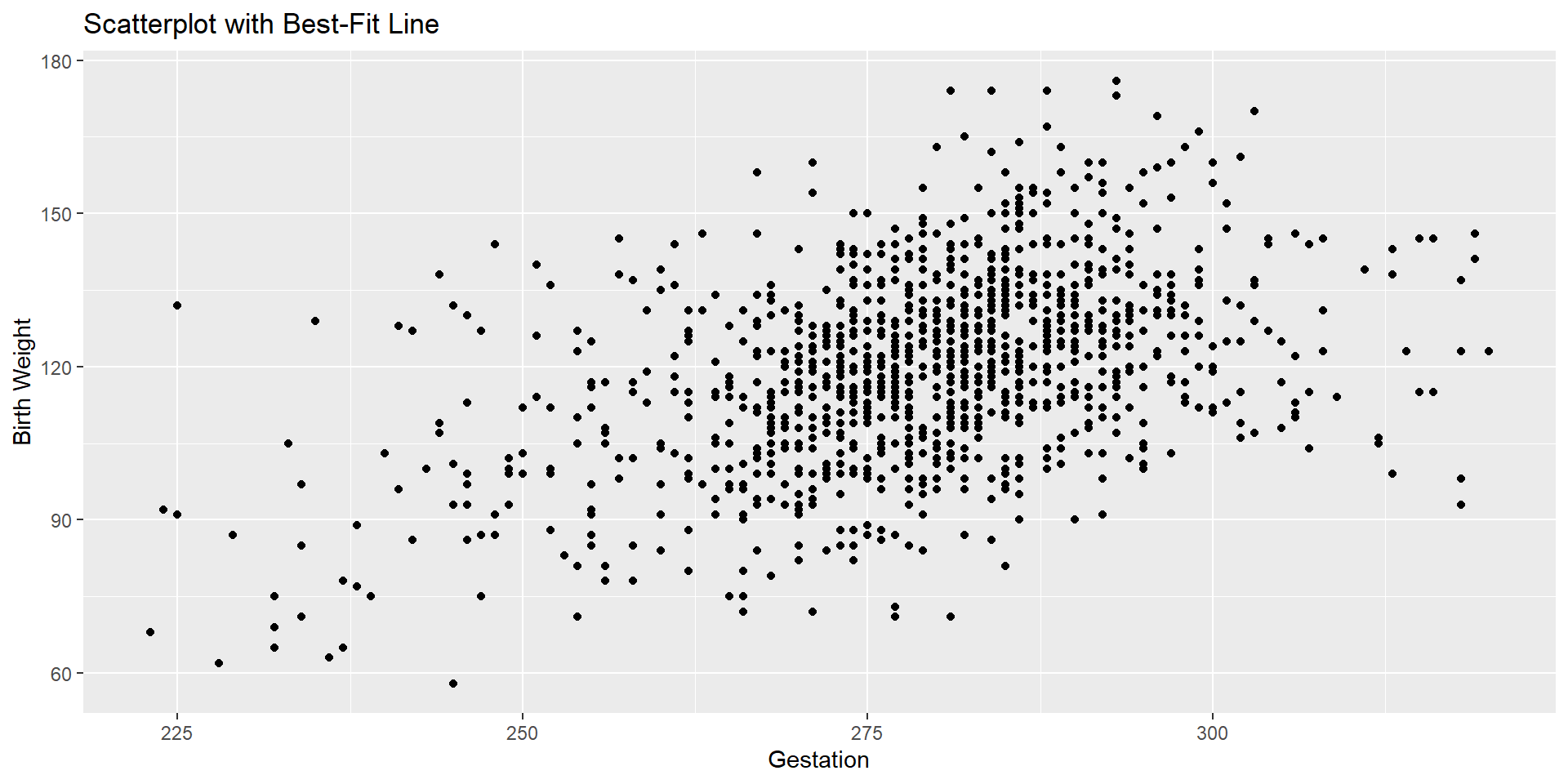
Let’s Brainstorm Together!
- Leverage points vs influential points
- Compare those correlation coefficients that we calculated
- Do we need to delete those outliers?
Bivariate Regression
Research Question and Data Set
Understanding Birth Weight and Gestation: A Least Squares Regression Analysis
In the area of prenatal care and childbirth, understanding the relationship between gestation period and birth weight is crucial. We often speculate about how the duration of pregnancy might be related with the weight of a newborn.
In the babies dataset, each observation includes information about gestation period and birth weight. We want to investigate if there’s a linear relationship between these two variables.
Hypothesis Testing
We will test the null hypothesis
\[ H_0: \beta_1 = 0 \] against the non-directional alternative
\[ H_A: \beta_1 \neq 0 \] Verbal Hypotheses are as follows:
\(H_0\): Birth weight is not linearly related to gestation
\(H_A\): Birth weight is linearly related to gestation
Important
Note that the test on \(\beta_1\) asks whether, assuming that the linear model holds, we can conclude that the slope is nonzero.
Checking Conditions/Assumptions:
Assumptions/Conditions
1. The observations should be independent
2. Linearity
- The relationship between the explanatory and the response variable should be linear.
- Check either using (1) a scatterplot of the data, or (2) a residuals plot.
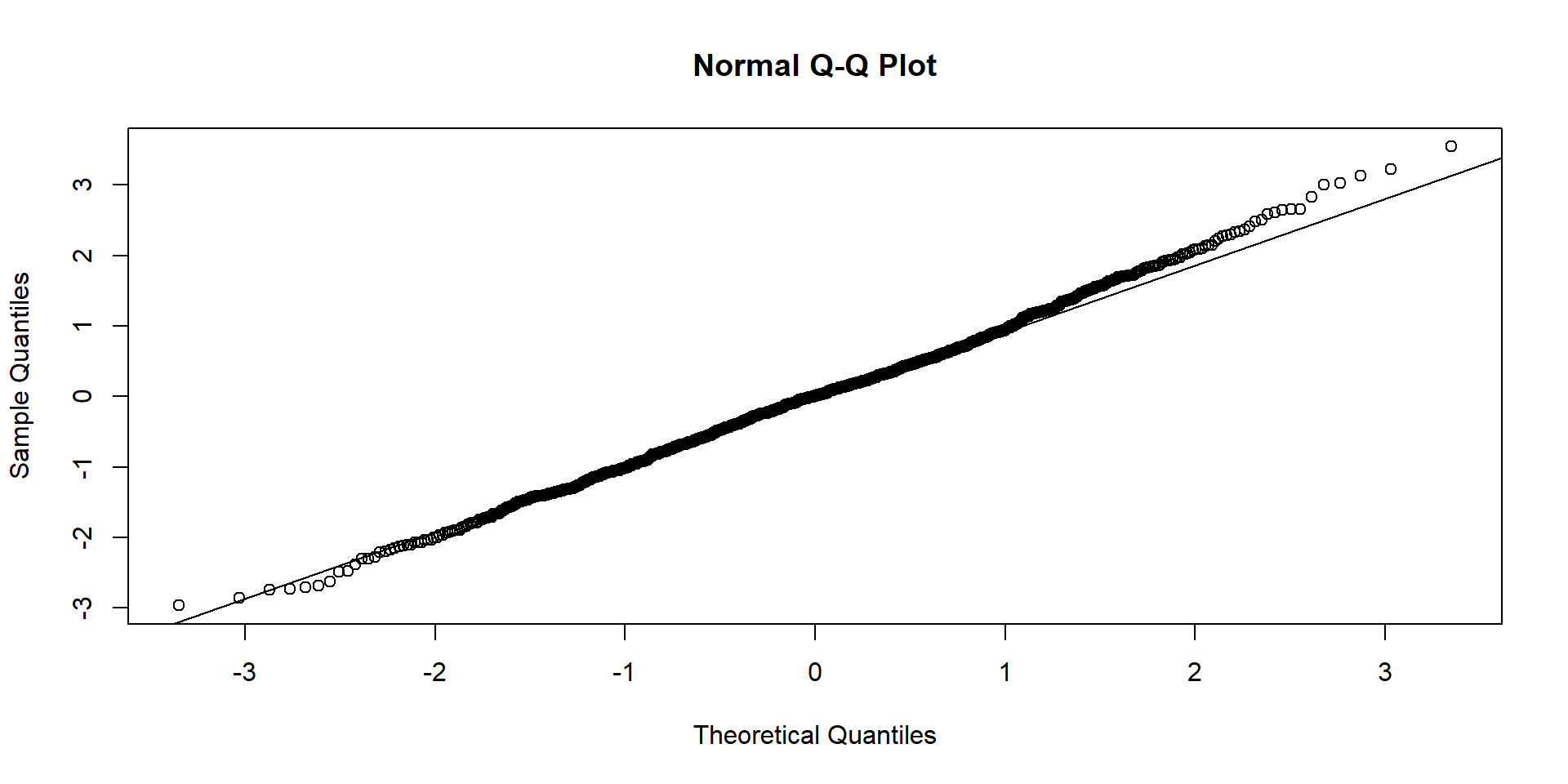
3. The residuals should be nearly normal.
This condition may not be satisfied when there are unusual observations that don’t follow the trend of the rest of the data.
Check using (1) a histogram or (2) normal probability plot of residuals.
4. Constant Variability - Homoscedasticity
The variability of points around the least squares line should be roughly constant.
This implies that the variability of residuals around the 0 line should be roughly constant as well.
Check using (1) a histogram or (2) normal probability plot of residuals.
Before Checking the Conditions
Step 2: Generate Your Model
IMPORTANT: The codes I use include the babies dataset. If you chose a different dataset, such as babies1 or babies2(see your environment pane), be sure to replace babies with the correct dataset name in all instances.
Before Checking the Conditions
- In bivariate regression, we need to first model the relationship and check the conditions after because we need residuals.
# I am using the babies dataset because I decided not to delete outliers
fit <- lm(babies$bwt ~ babies$gestation)
summary(fit)
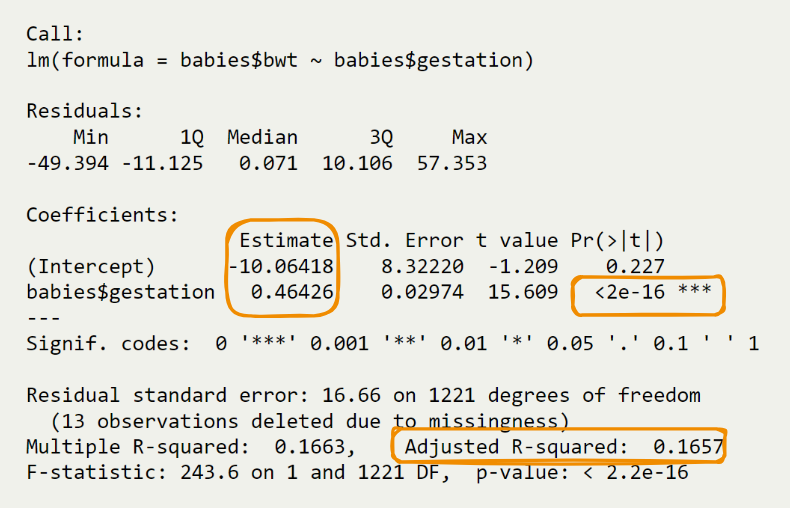
Call:
lm(formula = babies$bwt ~ babies$gestation)
Residuals:
Min 1Q Median 3Q Max
-49.394 -11.125 0.071 10.106 57.353
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -10.06418 8.32220 -1.209 0.227
babies$gestation 0.46426 0.02974 15.609 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 16.66 on 1221 degrees of freedom
(13 observations deleted due to missingness)
Multiple R-squared: 0.1663, Adjusted R-squared: 0.1657
F-statistic: 243.6 on 1 and 1221 DF, p-value: < 2.2e-16We employ the
lmfunction in R to perform a bivariate regression analysis.The formula
lm(babies$bwt ~ babies$gestation)indicates that we are modeling birth weight (bwt) as a linear function of gestation period (gestation).- The subsequent
summary(fit)provides detailed statistics about the regression model.
- The subsequent
Checking Conditions- Linearity with Scatterplot
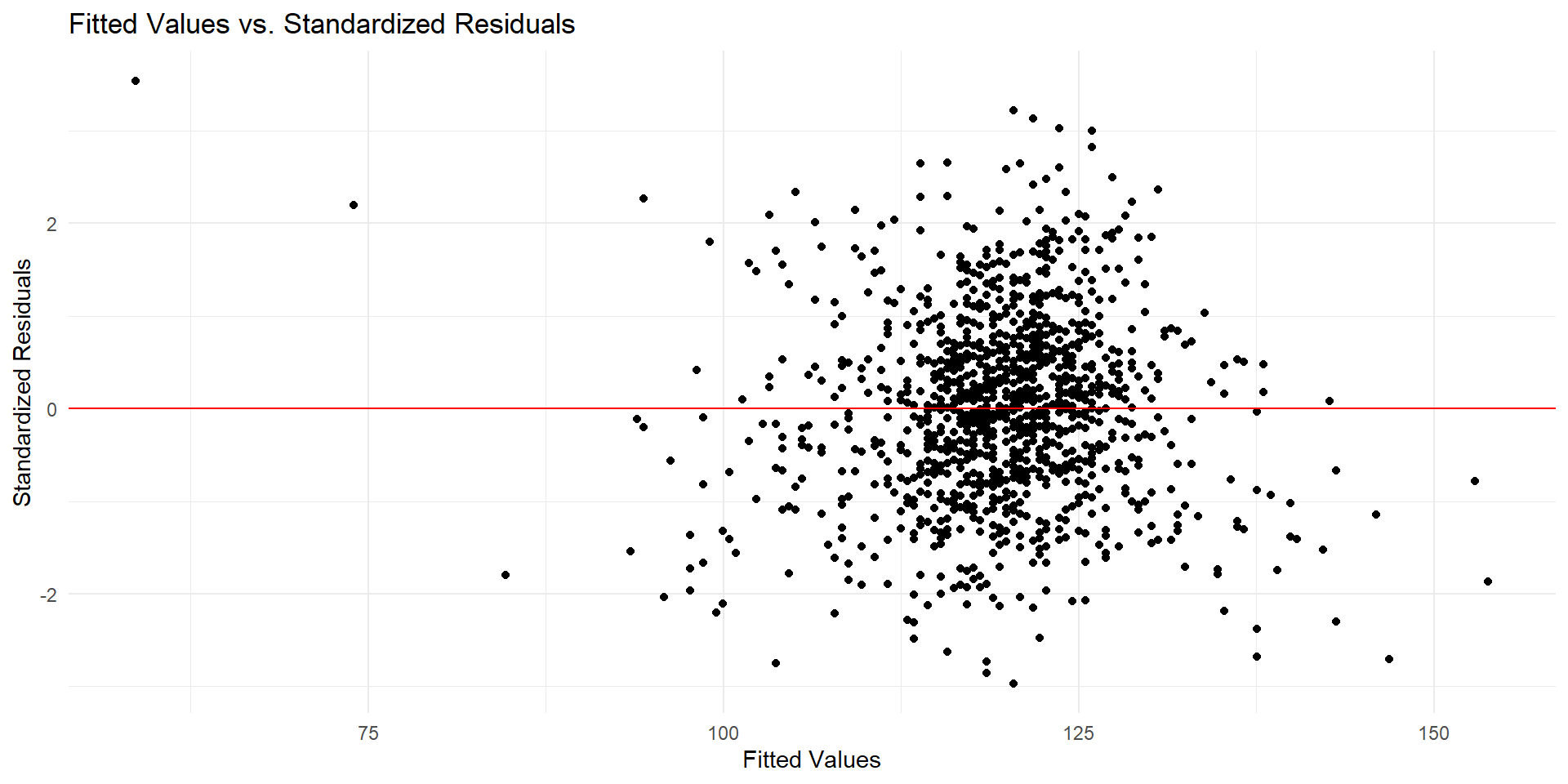
Checking Conditions - Linearity with Residuals Plot
# get list of residuals
res <- resid(fit)
res.stdres <- rstandard(fit) # standardized residuals
# produce residual vs. fitted plot
ggplot(data.frame(fitted = fitted(fit), stdres = res.stdres),
aes(x = fitted, y = stdres)) +
geom_point() +
geom_hline(yintercept = 0, linetype = "solid", color = "red") +
labs(title = "Fitted Values vs. Standardized Residuals",
x = "Fitted Values",
y = "Standardized Residuals") +
theme_minimal()Checking Conditions - Linearity with Residuals Plot
Checking Conditions - Normality
Checking Conditions - Homoscedasticity
# get list of residuals
res <- resid(fit)
res.stdres <- rstandard(fit) # standardized residuals
# produce residual vs. fitted plot
ggplot(data.frame(fitted = fitted(fit), stdres = res.stdres),
aes(x = fitted, y = stdres)) +
geom_point() +
geom_hline(yintercept = 0, linetype = "solid", color = "red") + # Add a horizontal line at 0
labs(title = "Fitted Values vs. Standardized Residuals",
x = "Fitted Values",
y = "Standardized Residuals")Checking Conditions - Homoscedasticity
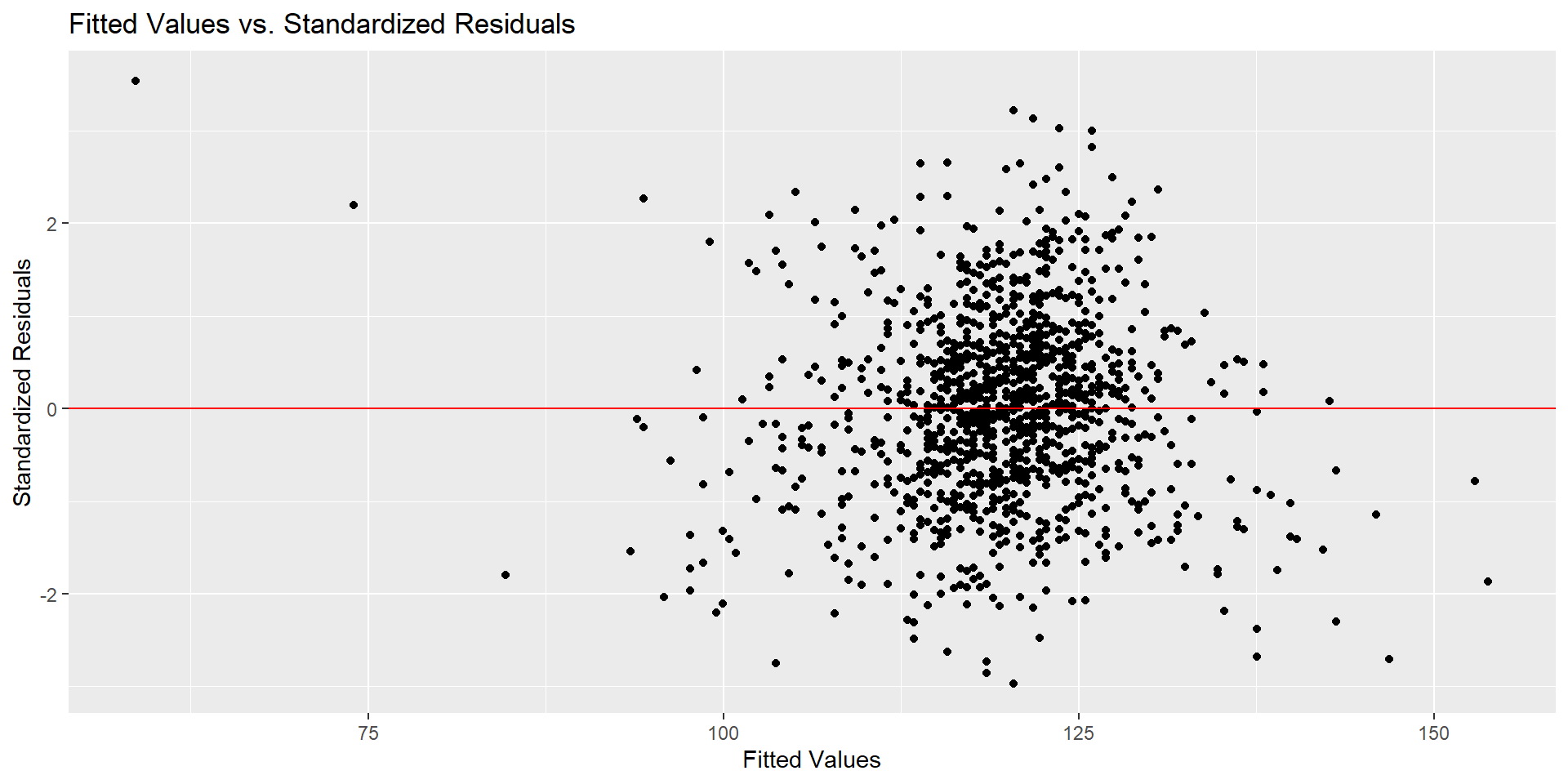
Draw Conclusion
Draw Conclusion
Call:
lm(formula = babies$bwt ~ babies$gestation)
Residuals:
Min 1Q Median 3Q Max
-49.394 -11.125 0.071 10.106 57.353
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -10.06418 8.32220 -1.209 0.227
babies$gestation 0.46426 0.02974 15.609 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 16.66 on 1221 degrees of freedom
(13 observations deleted due to missingness)
Multiple R-squared: 0.1663, Adjusted R-squared: 0.1657
F-statistic: 243.6 on 1 and 1221 DF, p-value: < 2.2e-16The regression equation is___________________
\[ \hat{y} = b_0 + b_1X \]